Hi
I try to reconstruct a SPECT image using et_demo_osem. I don't know if I am on the right track. Instead of loading activity.nii I load my SPECT-image of the Brain. My SPECT image consists of 128x128x384, in which only the first 128 out of the 384 is in the view with the correct energy. In other words the rest is scatter I guess.
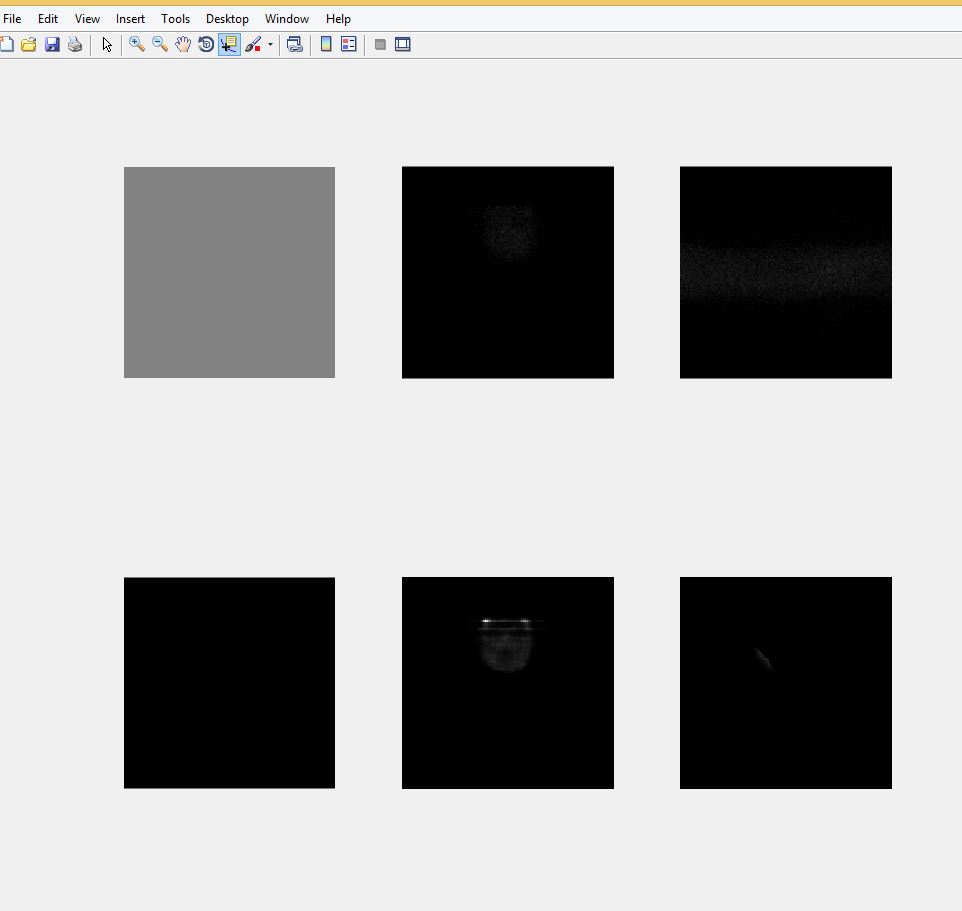
So far the code runs without errors but the reconstructed image does not look as I expected.
N = 128;
N_projections = 128;
cameras = linspace(0,2*pi,N_projections)';
%cameras = 2; The SPECT machine has two cameras?
psf = ones(5,5,N);
N_counts = 50e6;
iter_mlem = 50;
subset_order = 32;
GPU = 1;
phantom_type = 0; % 0 for 'brain FDG PET'; 1 for sphere in uniform background; 2 for spiral
%% Initialise plot
figure(1); subplot(2,3,1); image(zeros(N,N)); colormap gray; axis square off tight;
figure(1); subplot(2,3,2); image(zeros(N,N)); colormap gray; axis square off tight;
figure(1); subplot(2,3,3); image(zeros(N,N)); colormap gray; axis square off tight;
figure(1); subplot(2,3,4); image(zeros(N,N)); colormap gray; axis square off tight;
figure(1); subplot(2,3,5); image(zeros(N,N)); colormap gray; axis square off tight;
figure(1); subplot(2,3,6); image(zeros(N,N)); colormap gray; axis square off tight; pause(0.1);
%% Load or generate phantom
disp('Creating synthetic sinogram..');
%mask modifyed from placed in origo to placed so it fits the image
%mask = et_spherical_phantom(N,N,N,N*0.45,1,0,(N+1)/2,(N+1)/2,(N+1)/2);
mask = et_spherical_phantom(N,N,N,N*0.19,1,0,(N+1)/2,34,(N+1)/2);
if phantom_type == 0
phantom_file = 'BRAINTOMO.nii';
if not(exist(phantom_file,'file'))
fprintf('Synthetic data file %s cannot be found. \nPlease make sure that NiftyRec data is installed. \nIf NiftyRec was built and installed from the source code, \nrun CMake, enable the INCLUDE_DATA flag and run "make install".\n',phantom_file);
return;
end
phantom = et_load_nifti(phantom_file);
phantom1= double(phantom.img);
%my Brain image has 384 projections, however I only use the first 128
%because it is the activity with "correct" energy.
phantom2=phantom1(:,:,1:128);
phantom = phantom2 .* mask;
elseif phantom_type == 1
phantom = et_spherical_phantom(N,N,N,N/8,60,20,N/4,N/2,N/3) .* mask;
else
phantom = et_spiral_phantom(N,N,N*3,60,20,8,2,10,40,2) .* mask;
end
attenuation = 0;
figure(1); subplot(2,3,1); imagesc(squeeze(phantom(:,floor(N/2),:))); colormap gray; axis square off tight; pause(0.1);
%% Simulate SPECT scan
%I load my the sinogram (I use the same;
addpath('C:\Users\Christian Eschen\Dropbox\NiftyRec\niftyrec\matlab');
sinogram1=load_nii('C:\Users\Christian Eschen\Dropbox\NiftyRec\niftyrec\matlab\BRAINTOMO.nii');
sinogram=sinogram1.img;
sinogram=double(sinogram);
sinogram=sinogram(:,:,1:128);
%because 1:128 is the view with the right energy
disp('Visualising sinogram..');
for i=1:N_projections
figure(1); subplot(2,3,2); image(0.3*squeeze(sinogram(:,:,i))'); colormap gray; axis square off tight; pause(0.1);
if i<N/2
figure(1); subplot(2,3,3); image(0.3*squeeze(sinogram(:,i,:))); colormap gray; axis square off tight; pause(0.1);
end
end
%% Reconstruction:
disp('Reconstructing..');
activity = ones(N,N,N);
for i=1:iter_mlem
fprintf('\nMLEM step: %d',i);
activity = et_osmapem_step(subset_order, activity, sinogram, cameras, attenuation, psf, 0, 0, GPU, 0, 0.0001);
scale = 400;
figure(1); subplot(2,3,4); image(scale*squeeze(mask(:,floor(N/2),:).*activity(:,floor(N/2),:))); colormap gray; axis square off tight;
figure(1); subplot(2,3,5); image(scale*squeeze(mask(floor(N/2),:,:).*activity(floor(N/2),:,:))); colormap gray; axis square off tight;
figure(1); subplot(2,3,6); image(scale*squeeze(mask(:,:,floor(N/2)).*activity(:,:,floor(N/2))')); colormap gray; axis square off tight; pause(0.05);
end
%% Cleanup
if GPU
et_reset_gpu();
end
disp('MLEM Done');
I hope you can help
- Christian Eschen
|
 Author
Author
 Modifying et_demo_osem "Reconstruction of SPECT"
Modifying et_demo_osem "Reconstruction of SPECT"