Welcome GuestShow/Hide Header |
|---|
Welcome Guest, posting in this forum requires registration. |
|
|
Forum » NiftyRec » PET » Scaling of OSAMPEM with and without PSF
| Pages: [1] |
 Author Author |
Topic: Scaling of OSAMPEM with and without PSF |
|---|
| artm |
|
|||||||
| spedemon |
|
|||||||
| artm |
|
|||||||
| spedemon |
|
|||||||
| Pages: [1] |
Version: 1.0.34 ; Page loaded in: 0.104 seconds.
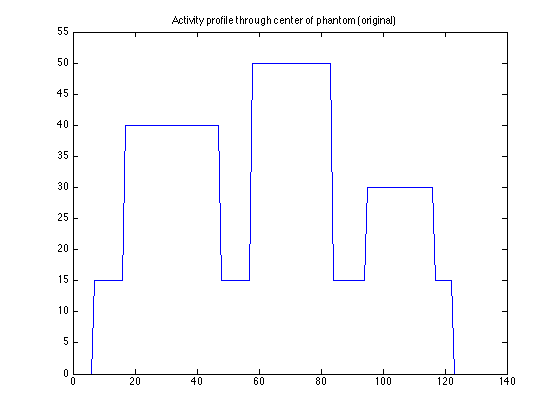
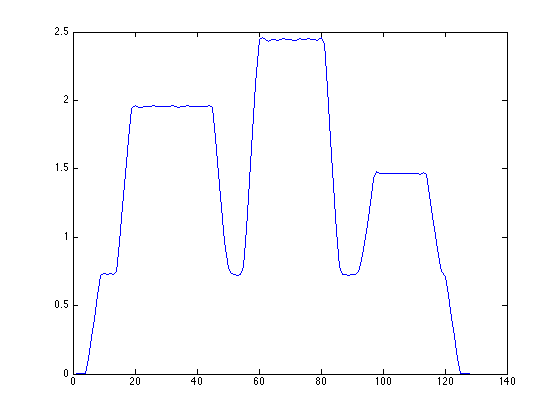
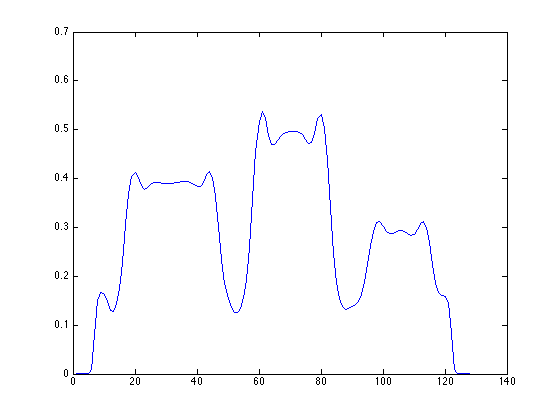
 Scaling of OSAMPEM with and without PSF
Scaling of OSAMPEM with and without PSF